Rootless Hair: DNA extraction & analysis for an alternative solution
December 16, 2019, Reading time: Less than a minute (134 words)

Recently developed, the InnoGenomics InnoXtract kit is formulated to increase recovery of small fragments of DNA from challenging samples. Following lysis, this bead-based extraction method produces DNA extracts that are free from inhibitors and capable of being used for multiple downstream applications. Preliminary studies have used extracts from the InnoXtract kit to reliably perform PCR amplification for use with both capillary electrophoresis (CE) and massively parallel sequencing.
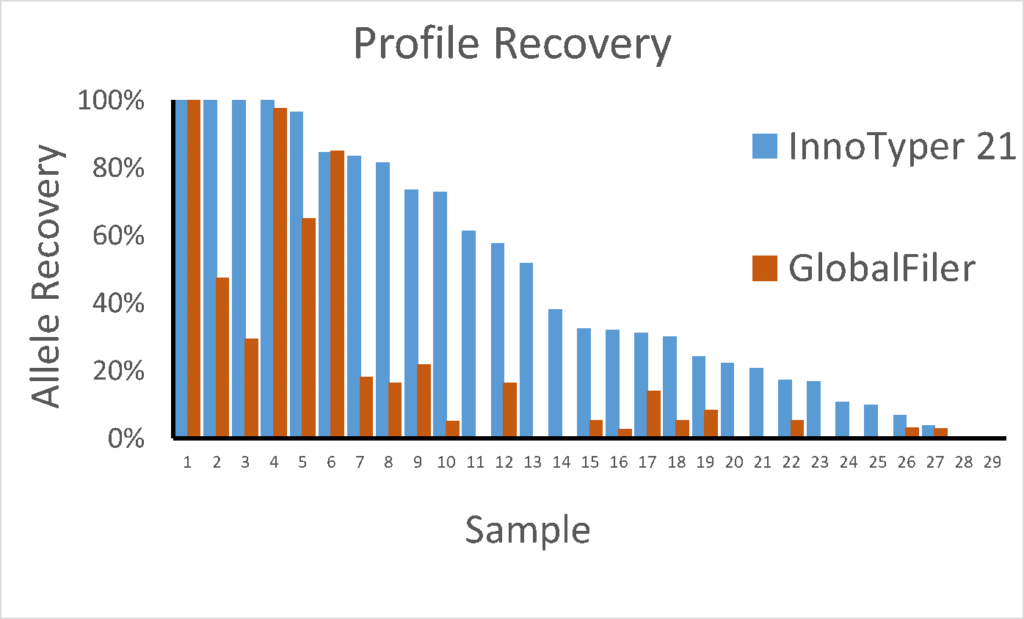
Rootless hair samples extracted with the InnoXtract chemistry have been used by researchers with the CE-based GlobalFiler, InnoTyper 21, and Big Dye Direct Sequencing Chemistry. Sample extracts were also amplified with the ForenSeq DNA Signature Prep Chemistry and Mitochondrial Control Region Solution for use with the MiSeq FGx.