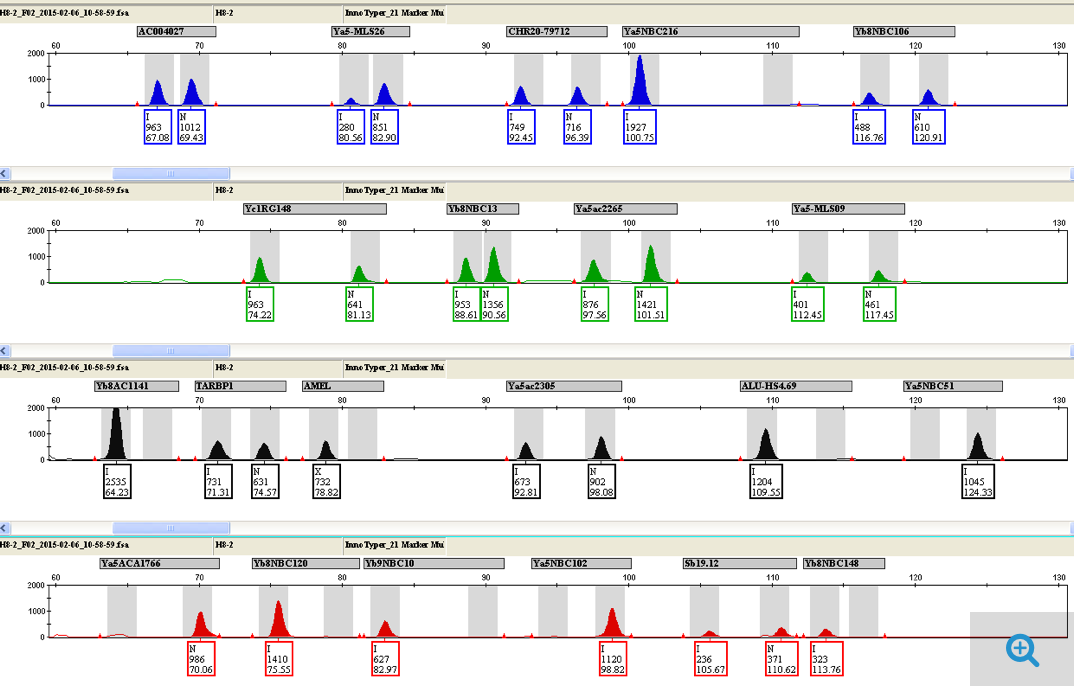
InnoTyper® 21
InnoTyper® 21
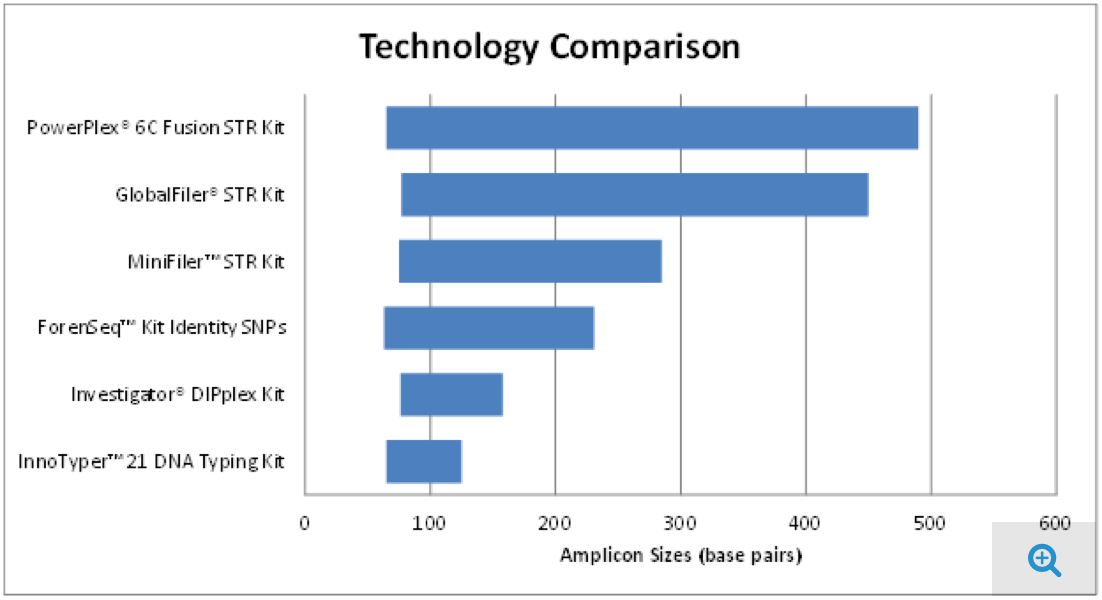
A small amplicon (~60-125 bp) DNA typing system for challenging forensic samples that is compatible with currently used Capillary Electrophoresis (CE) instrument platforms.
- Obtain DNA profiles from Rootless Hairs
- Achieve informative results with degraded/low level samples
- High sensitivity –obtaining profiles with as little as 15 – 30 picograms total DNA*
- Significantly more discriminating and less labor-intensive than mitochondrial DNA sequencing
- Compatible with DNA•VIEW DNA Analysis Software
This product is NOW AVAILABLE! Click here to try in your lab.
Order Products
Related Products
Product Details
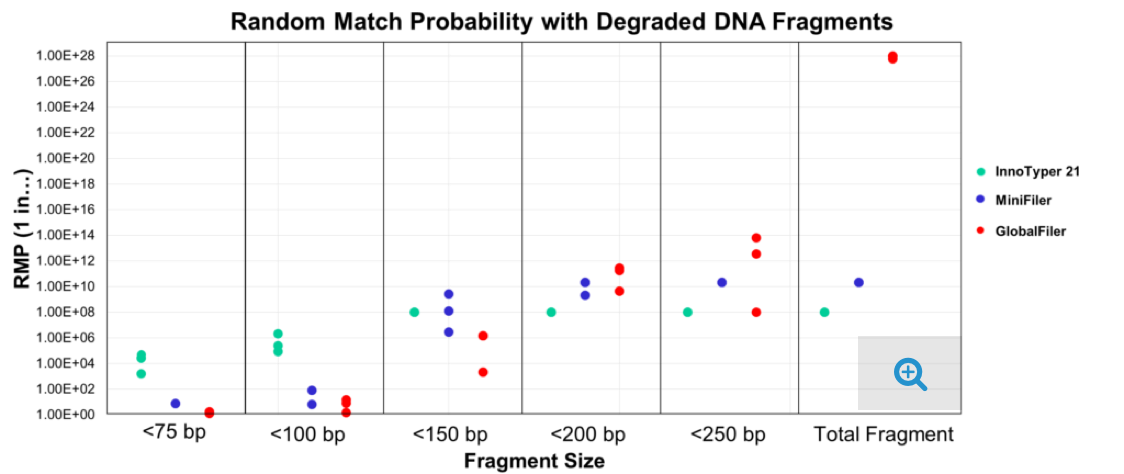
With the ability to obtain informative results from heavily degraded samples with as little as 15 – 30 picograms total DNA*, InnoTyper 21 provides a discriminating alternative to mitochondrial DNA (mtDNA) sequencing, which is currently the genotyping standard for degraded samples such as old human remains and hair shafts. InnoTyper 21 is significantly more discriminating and less labor-intensive than mtDNA sequencing, and does not require a specialized workflow.
InnoTyper 21 kit proved to be more discriminating than traditional STR multiplexes for DNA fragments less than 150 base pairs in size (For complete study details, see here.)
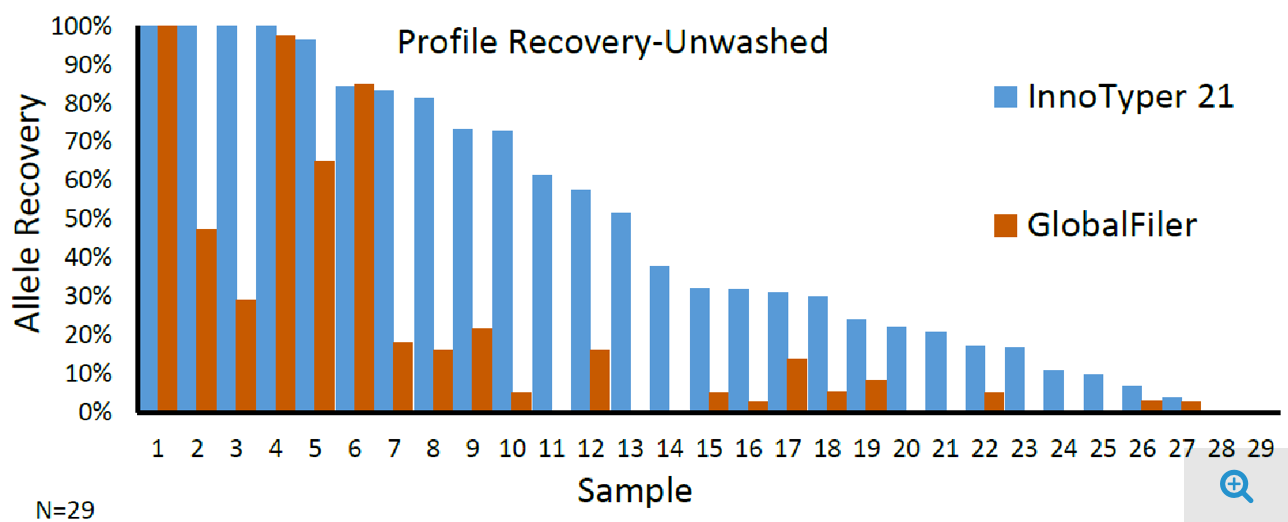
Studies have also demonstrated the utility of InnoTyper 21 for highly compromised human remains, i.e. bone fragments, that have failed to yield informative results using other methods, as shown in Figure 4. (For complete study details, see here.)
Resources
- Ryan Gutierrez’s AAFS presentation
- Developmental validation of InnoTyper 21, a nuclear DNA typing system based on retrotransposable element polymorphisms for degraded forensic samples (ISHI 2016 Poster)
- Validation of the InnoQuant and InnoTyper 21 Kits for Use on Forensic Samples (DNA Solutions ISHI 2016 Poster)